

(2005) novoSNP, a novel computational tool for sequence variation discovery, Genome Res 15, 436–442.
View chromatogram bioedit software#
(2005) SNPdetector: a software tool for sensitive and accurate SNP detection, PLoS Comput Biol 1, e53. A., Yakub, I., Wei, S., Sood, R., Rowe, W., Liu, P. (2003) Automated identification of single nucleotide polymorphisms from sequencing data, J Bioinform Comput Biol 1, 253–265.
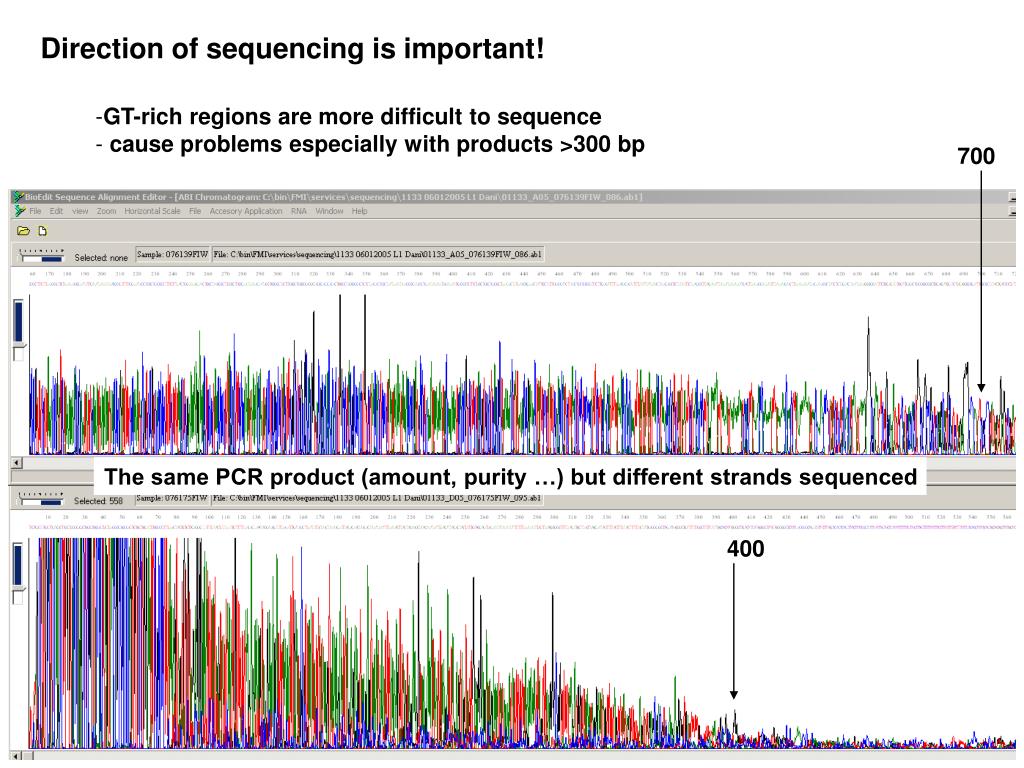
The sequences you are working with were prepared by theĭavidson lab from DNA fragments cloned in the pSTBlue-1 plasmid vector. Save the reverse complement as a text file Sequence will be at the end of the reverse complement. Sequence complementary to the beginning of your original unedited forward Note that this is also displayed in a 5'-3' direction, so the The sequence of the original template strand, the Reverse Complement mustĬlick on the view menu (for the original uneditedįile), and check Reverse Complement. The sequence present in the original file is the sequence of Preferably, your own disk or network account.) That sequences after 400-500 bases become increasingly unreliable, and are notĬlick on the File menu, Export as text. Is colour-coded to match the colour that one of the 4 bases is displayed Should be able to make a visual judgement about which base should be present Still see some “N”s in the sequence where the computer cannot make a call. The computer will already have called most of the bases from Sequence that can be edited without changing the original sequence. You should be able to clearly see the peaksĬlick on the view menu, and check editable Scale and Vertical scale bars to the top left of the image. Should recognise it as an ABI Autosequencer Trace file and open it as aĪdjust the size of the chromatogram trace with the Horizontal

(You may have to scroll down the programĬlick on File menu, Open. Also copy the file pstblue1vector.txt toĬlick on Start, Programs, and Bioedit. Each group should choose one of the sequenceįiles on the disk, and copy it from drive A to the desktop. There are 4 disks containing sequence files.
View chromatogram bioedit Pc#
This week, and the edited sequence files will be analyzed next week.Įditing (and most of the analysis) will be done usingīioEdit, a freeware sequence analysis program developed by Tom Hall at NorthĮach group should log on to a PC using the class ID bisc431 This weeks’ lab will be held in Room B8218 (the Biology PC


 0 kommentar(er)
0 kommentar(er)
